梅雅俊(Yaa-Jyuhn Meir)
|
Yaa-Jyuhu Meir |
||
|
Position |
Associate Professor |
|
|
Education |
Ph.D. in Cell Biology, Vanderbilt University, USA |
|
|
|
||
|
Office Tel |
+886-3-211-8800 ext.3641 |
|
|
Fax |
+886-3-211-8700 |
|
|
Laboratory |
Stem Cell & Molecular Genetics Laboratory |
|
|
Spciality |
Stem Cell Biology, Gene therapy, and Molecular Genetics |
|
|
Lab & Research Interest |
||
|
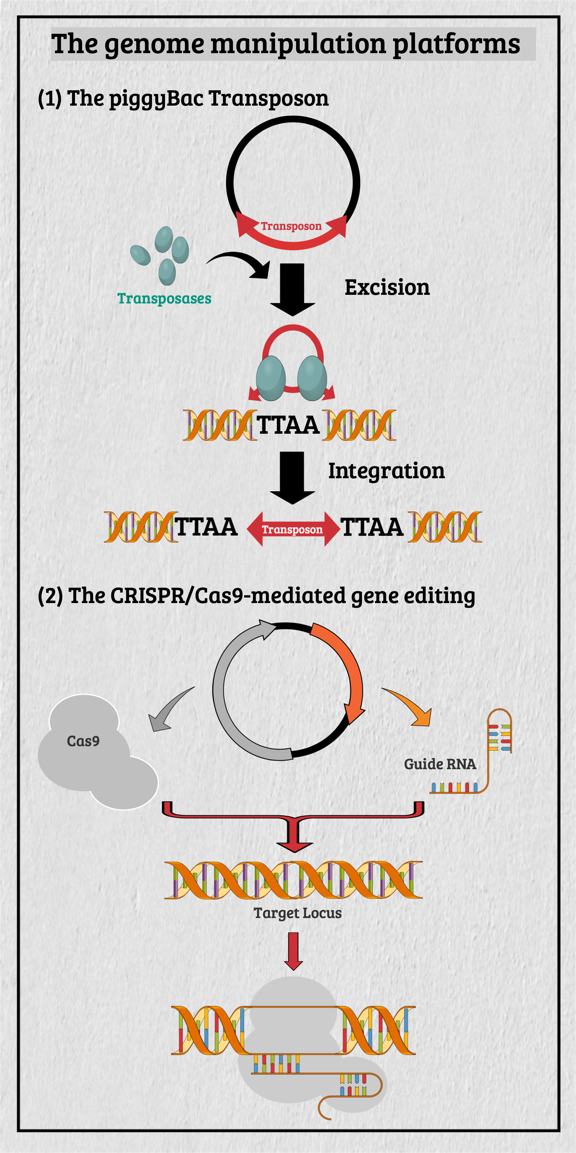
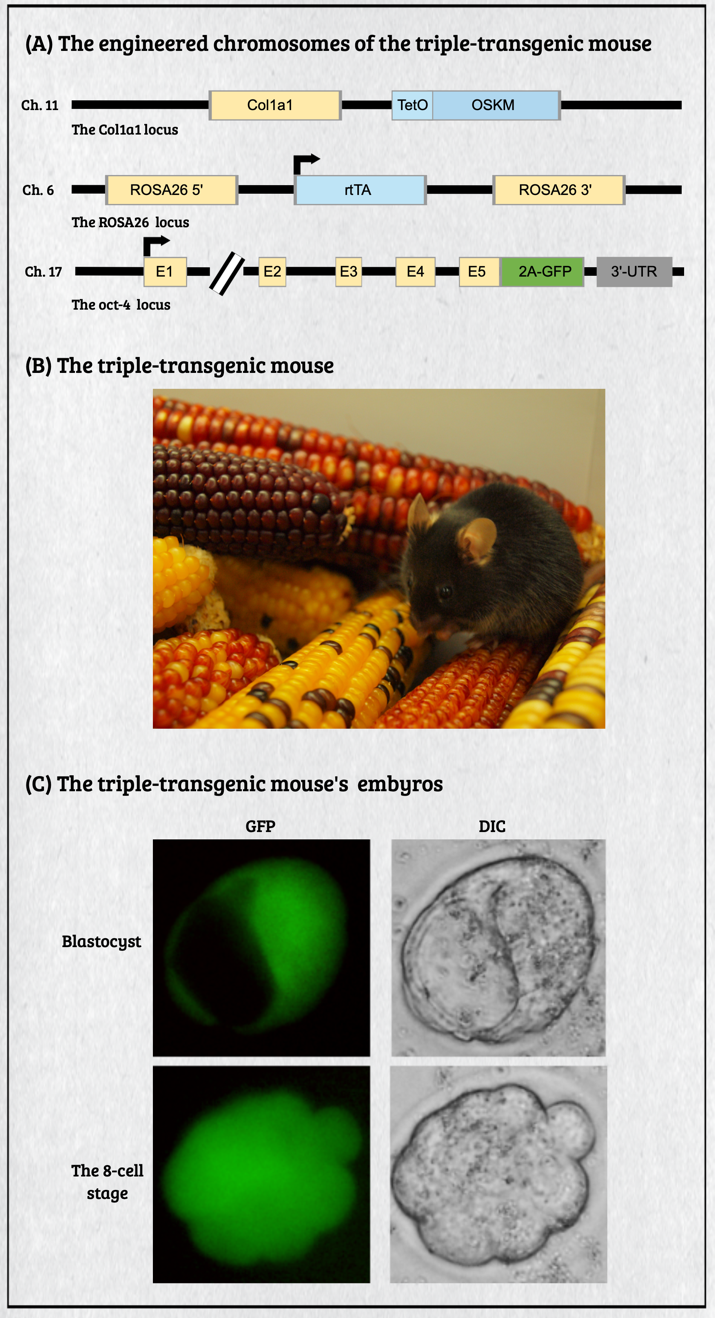
Multicellular organisms contain a particular group of cells, named stem cells, which possess the capacity for self-renewal and differentiation; in other words, stem cells serve as a reservoir of new cells to replace old or dying cells. With such remarkable capabilities, stem cells have long been considered as the "Fountain of Youth" for their ability to repair and replace lesions or aged tissues. They have been highlighted as the future direction of clinical sciences and a significant global bio-industry trend. This idea has been further boosted by the seminal finding of somatic reprogramming, which reverses the developmental timeline to restore stem cell characteristics from highly differentiated cells. The resulting cells, which possess many characteristics of embryonic stem cells (ESCs), are called induced pluripotent stem cells (iPSCs). This key finding provides a source of stem cells for cell replacement therapy and creates unprecedented opportunities for studying the development of organisms and diseases' etiology. The central interests of my lab are to study how cell fates are determined, how cells maintain their differentiated status, and which signals can alter cell identity to promote regenerative capability. Accordingly, my lab has focused on two primary directions: (1) unraveling the decision-making mechanisms of route choices during the pluripotency-acquiring process; (2) manipulating the genetic networks to generate the therapeutic progenitor cells of the specific tissues. In that vein, it relies heavily on a highly efficient genome manipulation platform for probing the dynamic molecular networks of pluripotency and detecting the consequent change of the cell fate. The versatile genome manipulation platforms Accordingly, my lab has established two genome manipulation platforms: the piggyBac transposon system and the CRISPR/Cas9-mediated gene-editing system. Transposons are one of the most popular genome manipulation tools that have been applied to the study of gene function in plants and invertebrates. Although 40% of the vertebrate genome comprises transposon-like elements, DNA transposons' mobilization has never been detected in mammals. Therefore, transposons have long been considered an ideal non-viral therapeutic vector system, harnessing to fix human genetic disorders with a large deletion on a chromosome. Works from our laboratory and others found that the piggyBac transposon, derived from the cabbage looper moth, can jump efficiently in different mammalian genomes even when carrying a large chunk of exogenous genes. Since then, the piggyBac transposon has gained its momentum for genome manipulation. It not only has high transposition efficiency in various mammalian cells, but it also has the unique property of being removed from the genome without a trace. Thus, the piggyBac transposon-based vector system has been considered one of the best therapeutic vehicles for gene therapy because of avoiding the "genome pollution" or an immune response as seen with viral vectors. My laboratory has currently advanced the piggyBac transposon-based genome manipulation system to manipulate the mammalian genome's structure and function efficiently. Also, we included the CRISPR/Cas9 system in applying its robust gene editing function on creating an isogenic cell with different single nucleotide polymorphisms (SNPs) in iPSCs for disease modeling. These well-established genome manipulation platforms allow us to unravel genomic plasticity by detecting chromatin remodeling's dynamic process during differentiation, somatic reprogramming, and regeneration. Adopting somatic reprogramming to study the genome plasticity When a highly differentiated cell acquires stem cell characteristics, the reversal process of cell differentiation allows us to study the genome's plasticity during somatic reprogramming. However, the current technology for creating iPSCs by exogenously introducing Yamanaka factors results in a heterogeneous population, which obscures the genetic pathways governing the dynamic process of somatic reprogramming. To overcome this difficulty, an inducible triple transgenic "The Col1a1 4F2A Oct4-GFP triple transgenic mouse" was created with the following genomic modifications: (1) a constitutive rtTA expression from the ROSA26 locus; (2) a pluripotent marker Oct4-ires-GFP knocked-in to the endogenous Oct4 locus; and (3) tetO-regulated iPSC quartet factors (Oct4-Sox2-Klf4-cMyc; OSKM) knocked-in to the Col1a1 locus. The exact genetic pathway that allows each cell type derived from this animal to regain pluripotency may directly relate to the cell's origin and its differentiation status. By creating individual clones, each representing an independent reprogramming event, we can identify factors that directly cause a specific phenotype (e.g., accelerating or preventing iPSC formation), which might otherwise be obscured by the heterogeneous cell population. Cellular reprogramming is a multifactorial and multistep procedure during which several fundamental cellular processes are coordinated sequentially until the pluripotent state is reached. Although iPSCs and nuclear transplantation are artificial ways of acquiring stem cell characteristics, there is increasing evidence from model animal studies that somatic reprogramming also occurs in vivo to replenish aged and damaged cells to maintain tissue homeostasis. Perhaps some cells inappropriately gain the potency of cell proliferation and consequently become tumor cells through a similar dedifferentiation process as seen in iPSC formation. Therefore, genetically dissecting the reprogramming process will provide a more in-depth insight into how programming factors or ambient stimuli reverse normal development and whether the mechanisms that usually prevent reprogramming in vivo may also involve in tumorigenesis. Also, identifying the different players involved in regaining pluripotency in distinct cell types will allow us to elucidate the principles of mammalian development and the mechanisms that usually prevent cells from undergoing malignant transformation. Ultimately, these studies may generate the desired cell state from existing cell types for cell replacement therapy and promote drug development in an individual setting.
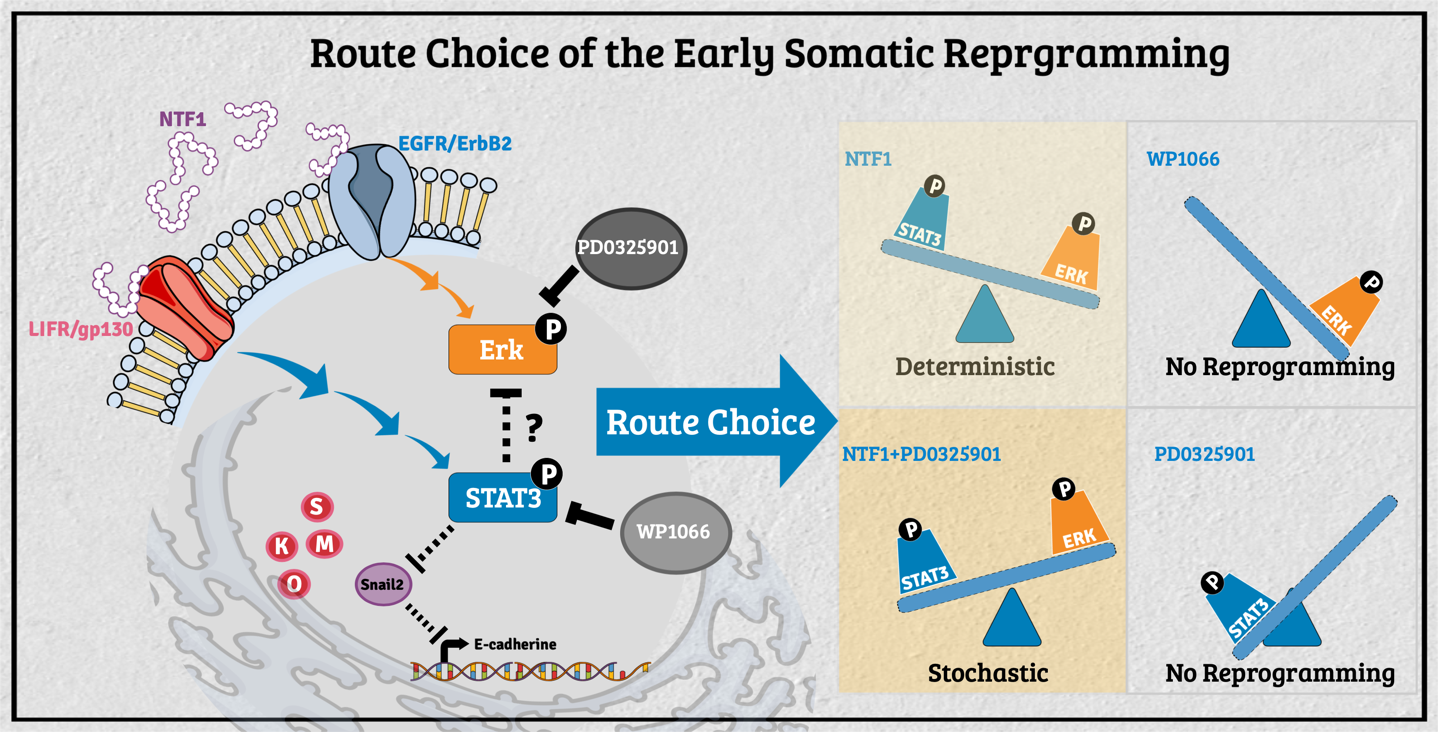
The decision-making of the reprogramming route choice affected by its residing microenvironment In the past, researchers have mainly explored the intrinsic capability of stem cells for tissue replacement and regeneration by unraveling the mechanisms underlying self-renewal and the maintenance of pluripotency. A growing body of evidence in different model organisms indicates that stem cells' behavior is critically influenced by their neighboring cells/tissues. In that regard, stem cell fate's dynamic status is tightly controlled through the concerted action of stem cell-intrinsic factors and signals derived from the milieu or niche in which the stem cell resides. The stem cell niche serves as a functional unit for development and regeneration in many tissues. It also nourishes resident stem cells and provides local or systemic instructions for immediate response to the body's needs. Thus, decisions about a stem cell's fate depend on the neighboring tissues, extrinsic signals, and intrinsic properties. This context-dependent combinatorial complexity of the stem cell niche poses an enormous challenge to studying stem cell biology's underlying mechanisms. To unravel the interplay of signals derived from intrinsic and extrinsic factors, we need to reconstruct the stem cell niche in the in vitro settings that recapitulate the interplaying dynamic features between stem cells and their surrounding environment. To that end, we had engineered the feeder cells to provide different factors to observe the extrinsic effect during the process of somatic reprogramming. Previously, several modes of reprogramming have been reported in various cell types during iPSC induction. The molecular mechanism regarding the selection of different action modes, however, is still mostly unknown. We recently found the molecular events participating in such processes' decision-making at the onset of somatic reprogramming. The activity of STAT3 versus Erk1/2 reversibly determines the reprogramming mode entered; a lower activity ratio favors the deterministic process and vice versa. Additionally, extraneous E-cadherin facilitates the early events of somatic reprogramming, potentially by stabilizing the LIF/gp130 and EGFR/ErbB2 complexes to promote entry into the deterministic process. Our current findings demonstrated that manipulating the pSTAT3/pErk1/2 activity ratio in the surrounding milieu can drive different modes of action toward either the deterministic or the stochastic process in the context of OSKM-mediated somatic reprogramming. These findings will allow us to navigate an efficient cell fate manipulation strategy according to the needs and, in turn, to optimize the human iPSC in vitro culture system from the regenerative medicine aspect.
Understanding the route choice of cell fate decision during the reprogramming process, we applied these findings to regeneration medicine to provide an autologous cell replacement therapy. Here, we picked the human corneal endothelial dystrophy as our first task for the therapeutic target.
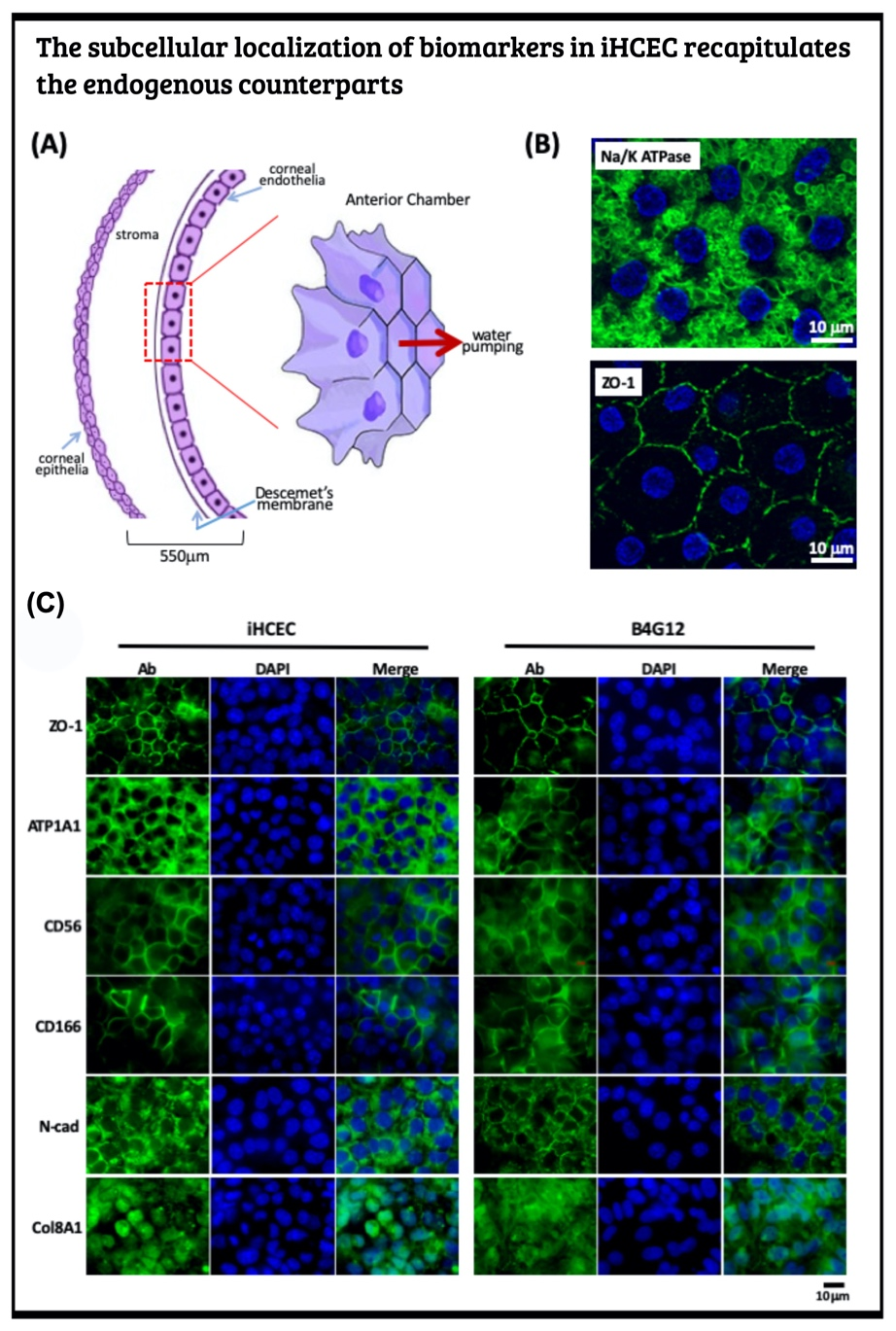
Generate human iPSC from PBMC and induce to the corneal endothelial cell via neural crest The human corneal endothelial cells (HCECs) constitute the innermost single cell layer of the cornea structure that governs corneal hydration's homeostasis. According to a global survey of corneal transplantation, 38% of 185,000 individuals who received corneal transplants each year suffer from corneal endothelium dysfunction. Due to the scarcity of donated corneas, only one cornea is available per 70 patients who need corneal transplantation. Instead of corneal transplantation, a recent therapeutic modality for the corneal endothelium dysfunction replaced the patient's corneal endothelial cells by the in vitro expanded primary HECEs. This novel strategy can compromise the shortage of corneal donation worldwide. Yet, it is a well-known fact that human corneal endothelial cells can no longer divide after birth and display only marginal proliferative activity in vitro. To overcome such hindrance, one shall identify the human corneal endothelial progenitors (HCEPs) instead of providing sufficient HCECs. In this regard, we first have to reveal the cell identity of bona fide HCEP from the anatomical structure and the corneal endothelial layer's developmental origin.
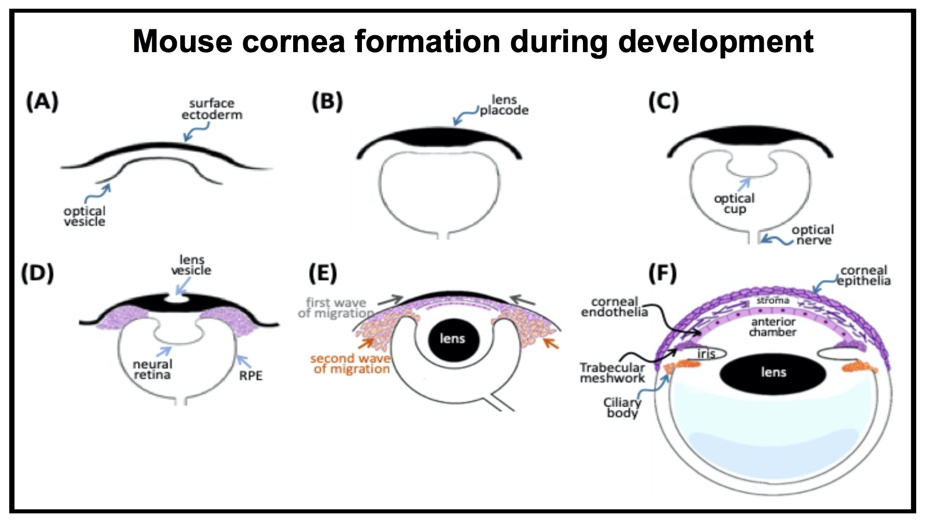
Cornea formation during development During vertebrate eye development, the future cornea is derived from two waves of mesenchymal cell migration. The first wave of mesenchymal cells migrates and fills the space between the surface ectoderm and the lens epithelium. These mesenchymal cells form a flat stratified layer separated by the extracellular matrix. The innermost stratified layer of mesenchymal cells creates an apicolateral connection with neighbor cells via tight junction structures. Thus, the endothelial monolayer of the cornea is formed. During the cornea and lens separation process, the second wave of mesenchymal cell migration occurs at the anterior eye. These cells appear at the future iridocorneal angle where the cornea meets the optic cup's anterior edge and form the future stroma of the iris and ciliary body. Shortly after creating the iris and ciliary body, the mass of mesenchymal cells occupying the iridocorneal angle will produce a trabecular meshwork and Schlemm's canal to complete the anterior eye's development.
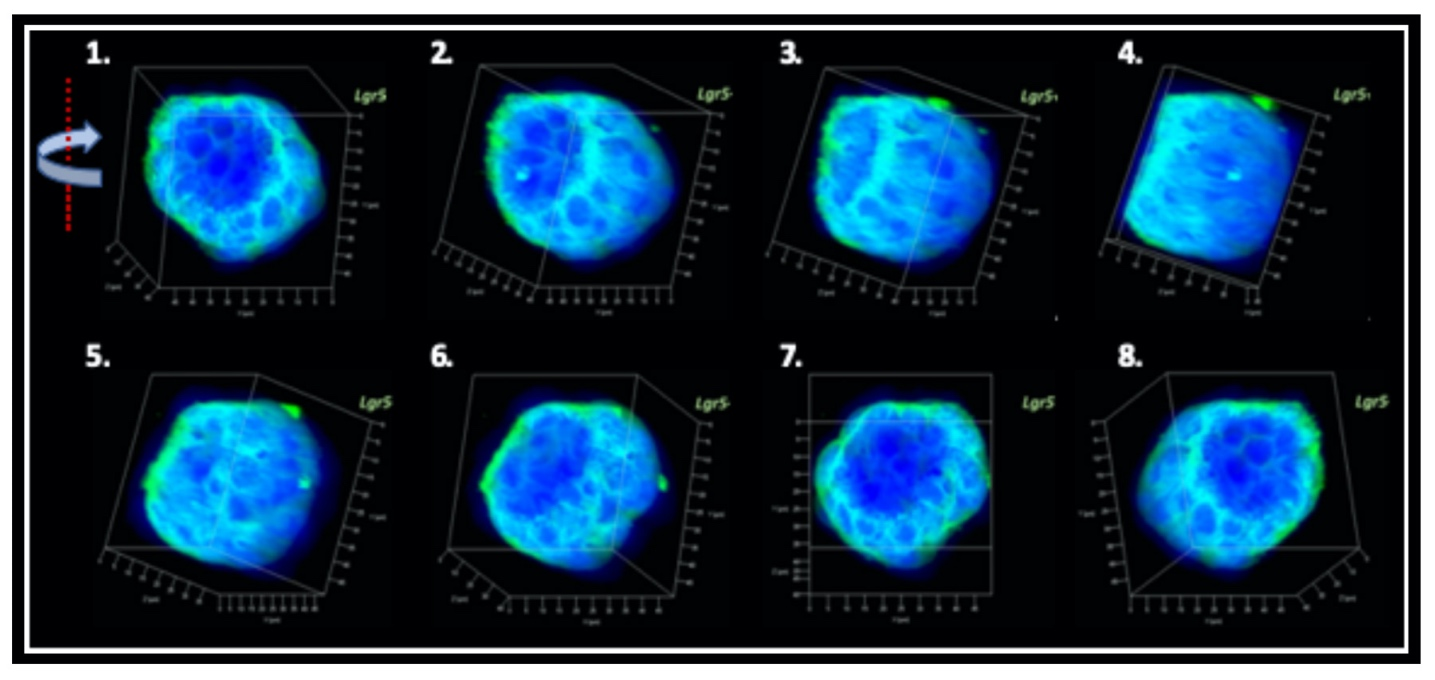
Producing the hiPSC-derived iHCEPs Prior reports recognized several potential corneal endothelial progenitor niches in the cornea, including the peripheral part of the endothelial sheet, transition zone, and trabecular meshwork. Although those cells within each region may possess different characteristics concerning their expression profiles, they can differentiate into the mature corneal endothelial cells. Nevertheless, these progenitor-like states cannot be captured and maintained in the cultural environment. Thereof, we set out to generate the induced human corneal endothelial progenitor (HCEP) from human iPSC. As mentioned above, corneal endothelial cells' origin came from the first wave migration of mesenchymal and neural crest cells during eye development. Therefore, we followed the cornea developmental path to generate the induced HCEPs (iHCEPs) by creating the neural crest fate first. As shown in the above figure, the iHCEPs resembled the endogenous HCEC counterpart in each gene expression and subcellular localization aspect. Generating the corneal endothelial organoid recapitulates the endogenous endothelial sheet To demonstrate the capability of being a progenitor cell, we placed the iHCEPs in a 3D culture to mimic the endogenous situation for producing the self-organized organoid of the corneal endothelial sheet. The figure below showed the confocal animation images of the iHCEP organoid in a 3D Matrigel-based culture system. The iHCEP self-organized organoid in the Matrigel-based 3D culture system forms a pouch-like configuration. The Lgr5, a progenitor marker, immunostaining (shown in green) depicted its expression around the iHCEC-organoid pouch opening (the counterpart of the endothelial layer's peripheral region) and gradually diminished toward the bottom (the endothelial layer's central area) of the pouch-like structure. The anatomical arrangement of progenitors in the pouch opening, but not in the base region, recapitulates the human corneal endothelial layer's architecture. We currently adopted the rabbits' cornea system to evaluate the therapeutic efficacy of our iHCEP and the iHCEP-derived organoid.
Closing Remarks The phenomenal somatic reprogramming provides new opportunities to understand the genome's plasticity, creates novel platforms for bio-industrial drug screening, and offers the therapeutic cells for regenerative medicine. Since it has been shown that iPSCs can model diseases and now we further generated organoids in a dish, there is an unprecedented opportunity to perform a high-throughput in vitro drug screening where the extent to validate the compounds' specificity and efficacy in a tissue & organ manner. |
||
|
Publications |
||
Peer-reviewed Publications (in chronological order)(1) JOURNAL
(2) POPULAR SCIENCE科學人 『跳躍基因活躍中』 NO.123. p40 (2012). 作者: 梅雅俊 教授 (3) PATENTSUS UTILITY PATENT US PROVISIONAL PATENT
|
||